
Welcome to ZooMS!
This page is for people who are new to ZooMS and would like assistance getting started. Before beginning, there are several primers and reviews available that explain the basic principles of ZooMS. We recommend reading these before proceeding further:
- Richter KK, Codlin MC, Seabrook M, Warinner C. 2022. A primer for ZooMS applications in archaeology. Proceedings of the National Academy of Sciences 119(20):e2109323119.
- Buckley M. 2018. Zooarchaeology by mass spectrometry (ZooMS) collagen fingerprinting for the species identification of archaeological bone fragments in Zooarchaeology in Practice: Case Studies in Methodology and Interpretation in Archaeofaunal Analysis, edited by Giovas CM and LeFebvre MJ. Springer International Publishing, pp. 227–247.
- Warinner C, Korzow Richter K, Collins MJ. 2022. Paleoproteomics. Chemical Reviews 122(16):13401-46.
- Kirby DP, Buckley M, Promise E, Trauger SA, Holdcraft TR. 2013. Identification of collagen-based materials in cultural heritage. Analyst 138(17):4849-58.
Collagen
ZooMS focuses on the peptide mass fingerprinting of type I collagen (COL1), a large triple helical protein found in a wide variety of animal tissues. At the molecular level, COL1 consists of a triple helix made up of three polypeptide α-chains (COL1A). In tetrapods, the triple helix is heterotrimeric, composed of two identical COL1A1 chains and one COL1A2 chain. In teleost fish, it is made up of three different chains (COL1A1, COL1A2, COL1A3), while a small number of species, such as the unicellular hydra, have homotrimeric COL1 composed of three COL1A1 chains.
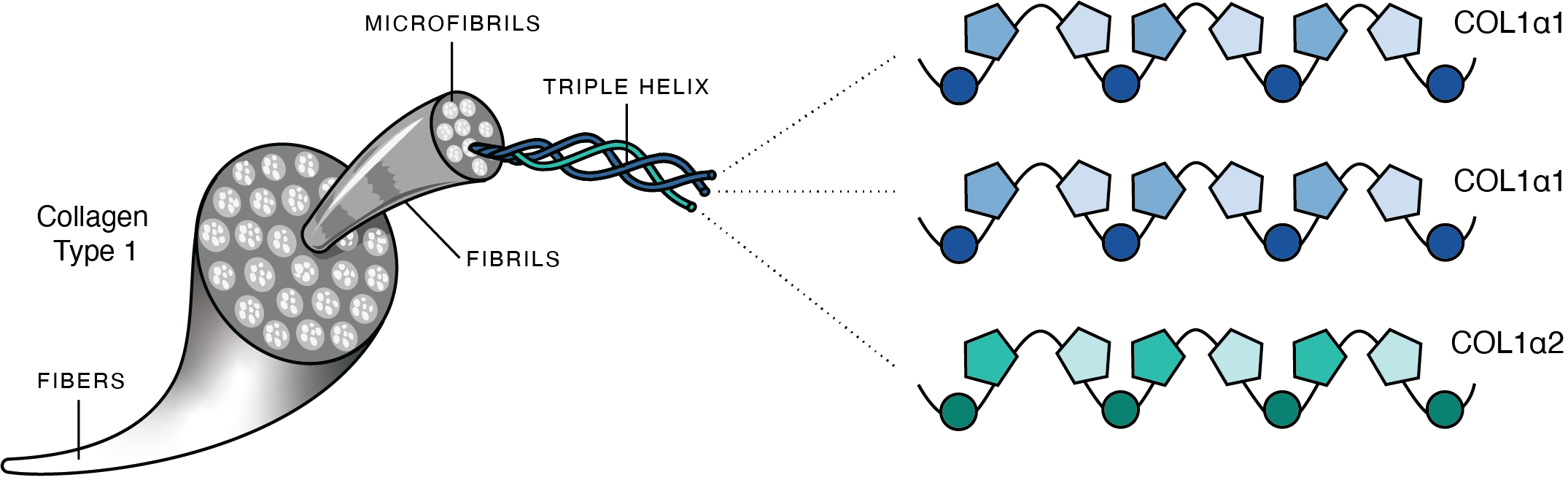 Schematic of mammalian collagen illustrating the hierarchical structure of type I collagen. Three COL1a chains twist together to form a triple helix. These then bundle together to form microfibrils, and microfibrils bundle together to form fibrils, and fibrils bundle together to form fibers. Image from Richter et al. 2022.
Schematic of mammalian collagen illustrating the hierarchical structure of type I collagen. Three COL1a chains twist together to form a triple helix. These then bundle together to form microfibrils, and microfibrils bundle together to form fibrils, and fibrils bundle together to form fibers. Image from Richter et al. 2022.
The amino acid sequence of COL1 is highly structurally and functionally constrained. Each chain consists of a repeating motif of G-X-Y with glycine (G), the smallest amino acid, fitting into the central core of the rotating triple helix. The remaining X and Y amino acid positions are disproportionately made up of proline and hydroxyproline, respectively, the latter being a posttranslational modification (PTM) of proline rarely found outside of collagens. Hydroxyprolines stabilize the triple helix through hydrogen bonding and can be always present (fixed modification) or variably present (variable modification) at a given amino acid position. Amino acids with bulky functional groups are almost entirely absent from COL1 because they disrupt or prevent the formation of the triple helix.
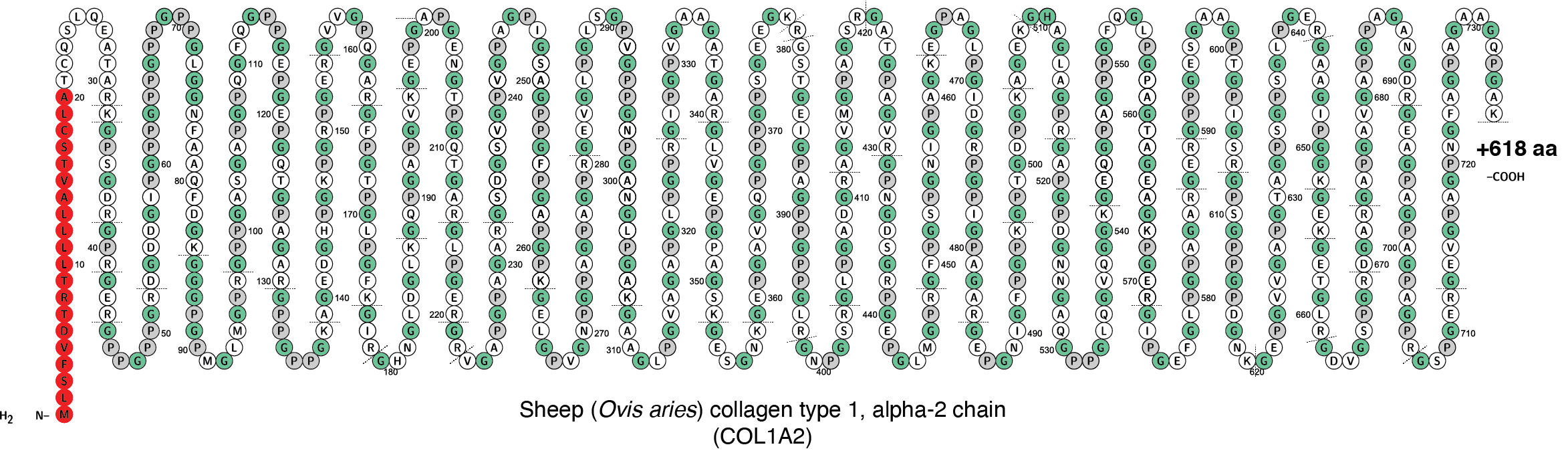 Schematic of the first 746 amino acids encoded by the sheep COL1a2 gene. The signal peptide, which is not present in mature collagen, is marked in red. The repeating G-X-Y motif has been highlighted by indicating each G in green and each P gray. Subsequent posttranslational modifications of some prolines into hydroxyproline are not shown. Tryptic cut sites are indiated by dotted lines. Image generated in Protter.
Schematic of the first 746 amino acids encoded by the sheep COL1a2 gene. The signal peptide, which is not present in mature collagen, is marked in red. The repeating G-X-Y motif has been highlighted by indicating each G in green and each P gray. Subsequent posttranslational modifications of some prolines into hydroxyproline are not shown. Tryptic cut sites are indiated by dotted lines. Image generated in Protter.
Because of the functional constraints of COL1, the protein evolves slowly. ZooMS analysis can typically resolve birds to the level of family or order; mammals to the level of genus or family; and fish to the level of species or genus. Differences in taxonomic resolution are related to differences in the proteins making up the collagen triple helix in these three groups, as well as differing functional constraints on collagen evolution related to body temperature and mechanical stress. See Richter et al. 2022 for a more detailed explanation of collagen structure, evolution, and taxonomic resolution.
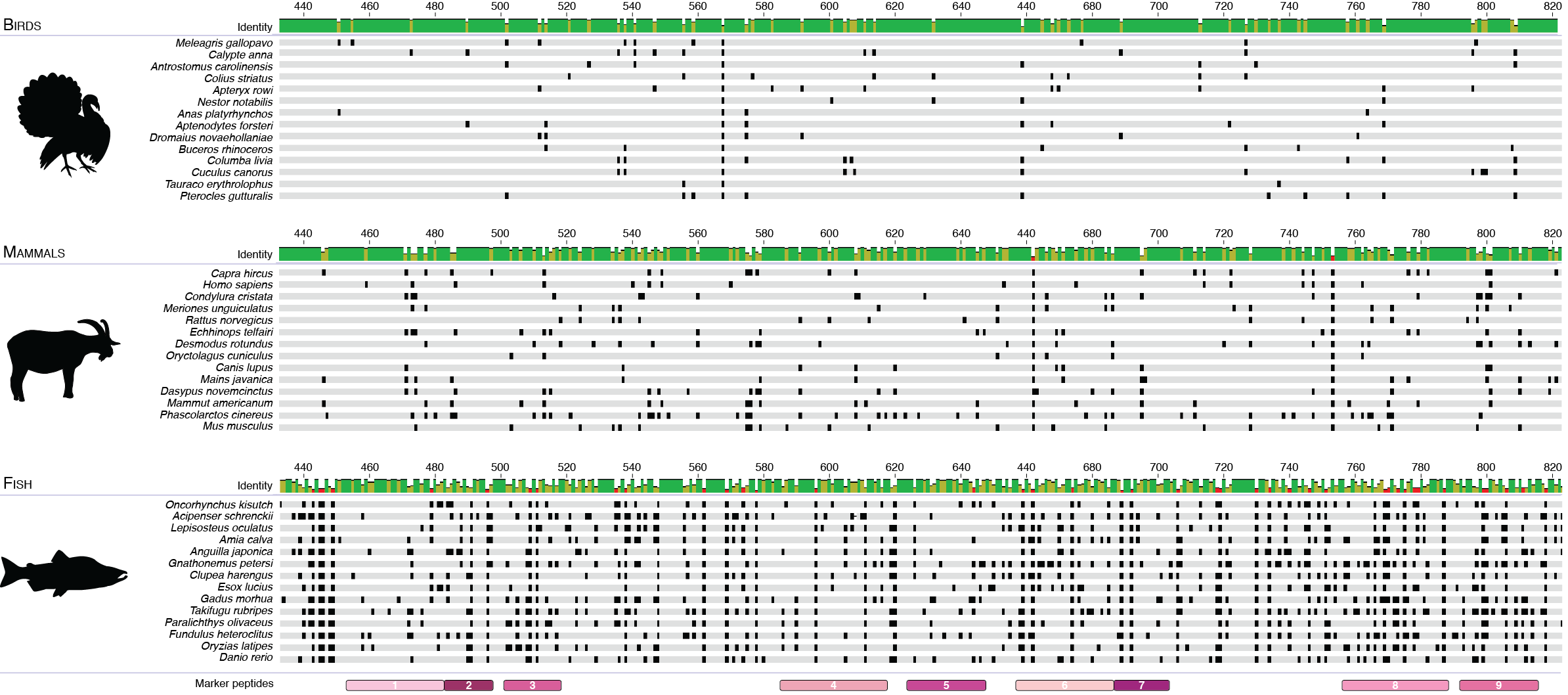 Alignment of amino acid positions positions 432-824 of the COL1ɑ2 protein from a diverse set of birds, mammals, and fish. Amino acid sequence differences from taxonomic group consensus are marked in black, and marker peptides are shown in pink. Fish have the highest sequence variation, followed by mammals, and then birds. Sequences were aligned using Geneious 2019.0.4 (Biomatters Ltd.). Image from Richter et al. 2022.
Alignment of amino acid positions positions 432-824 of the COL1ɑ2 protein from a diverse set of birds, mammals, and fish. Amino acid sequence differences from taxonomic group consensus are marked in black, and marker peptides are shown in pink. Fish have the highest sequence variation, followed by mammals, and then birds. Sequences were aligned using Geneious 2019.0.4 (Biomatters Ltd.). Image from Richter et al. 2022.
Mass spectrometry
ZooMS uses MALDI-TOF mass spectrometry to measure the mass-to-charge ratio (m/z) of individual collagen peptides. A typical trypsin digestion of COL1 produces many peptides, resulting in a characteristic “peptide mass fingerprint” for different taxa. MALDI-TOF stands for matrix-assisted laser desorption ionization (MALDI) time-of-flight (TOF) mass spectrometry. MALDI-TOF mass spectrometry is a three-step process whereby peptides that have been embedded within a matrix (typically α-cyano-4-hydroxycinnamic acid) are first vaporized and ionized using a laser, and then accelerated through a flight tube using electromagnets to separate the different peptides by size, and then finally sensed by a detector to determine each peptide’s mass. The data output is a mass spectrum, which is typically saved as an .xml file.
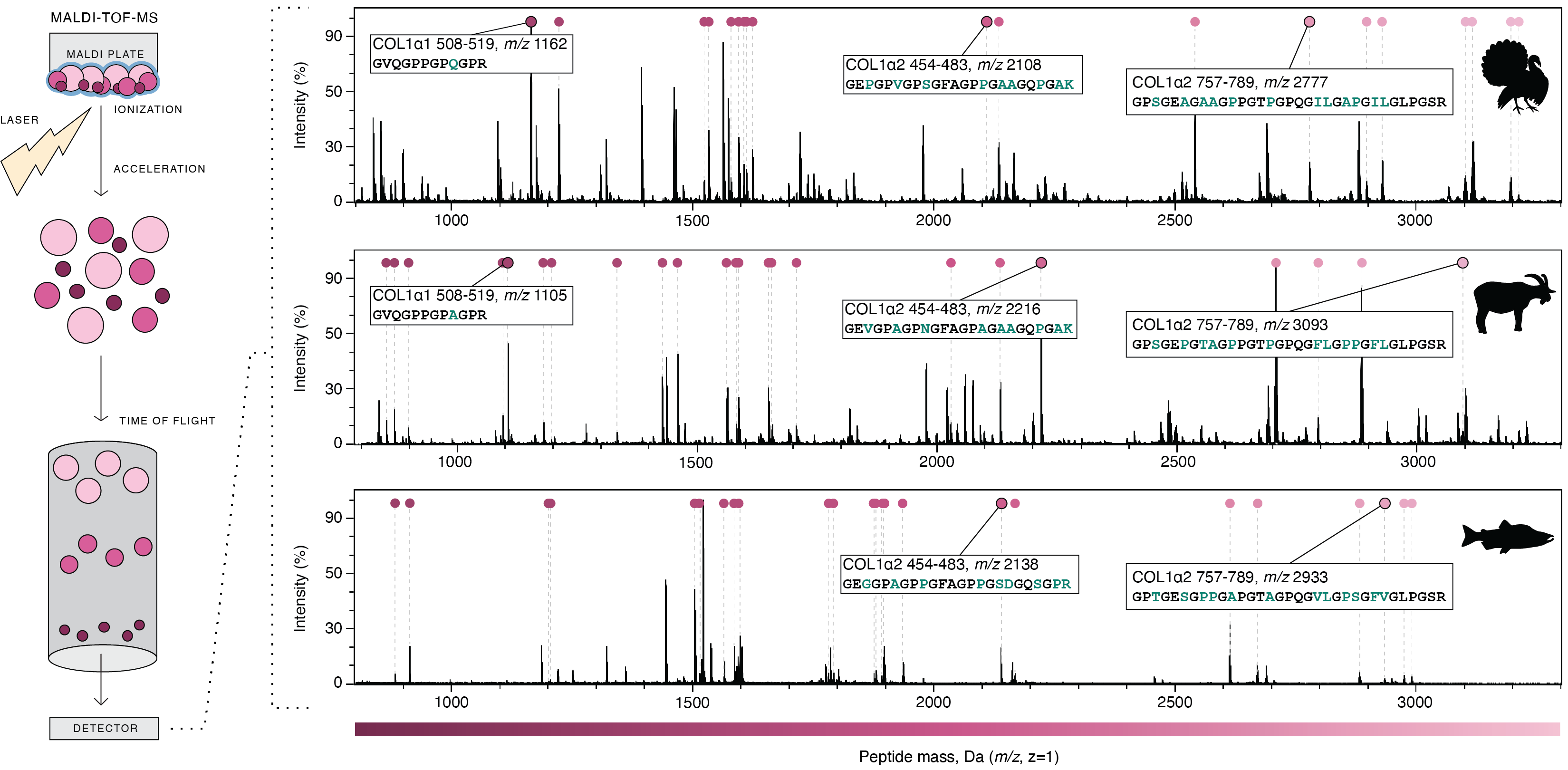 Steps of MALDI-TOF mass spectrometry and resulting COL1 mass spectra for turkey, goat, and coho salmon. Selected peptides that are useful for taxonomic disrimination are annotated. Image from Richter et al. 2022.
Steps of MALDI-TOF mass spectrometry and resulting COL1 mass spectra for turkey, goat, and coho salmon. Selected peptides that are useful for taxonomic disrimination are annotated. Image from Richter et al. 2022.
It is important to note that not all peaks present in a COL1 mass spectrum originate from collagen. Matrix peaks are also present, as are common laboratory contaminants such as keratins. These addional peaks must be exluded from analysis. Matrix peaks are typically low mass (<1,000 Da) and overlap with short collagen peptides. Problems may also arise with COL1 peaks. Peptides with the same or similar mass have overlapping peaks and may not be distinguishable, thus making them taxonomically unreliable markers. Peaks that are not specific because they can derive from different peptides also make poor marker peptides. COL1 also undergoes degradation through time, which can lead to peptide mass shifts, or even the loss of some peptides. Common chemical changes that are known to occur in archaeological collagen include deamidation of asparagine (N) and glutamine (Q) to aspartic acid (D) and glutamic acid (E), respectively. Each deamidation cause a +1 Da mass shift due to replacement of the amide with a carboxyl functional group. High-mass peptides are often underrepresented in archaeological remains due to poor preservation.
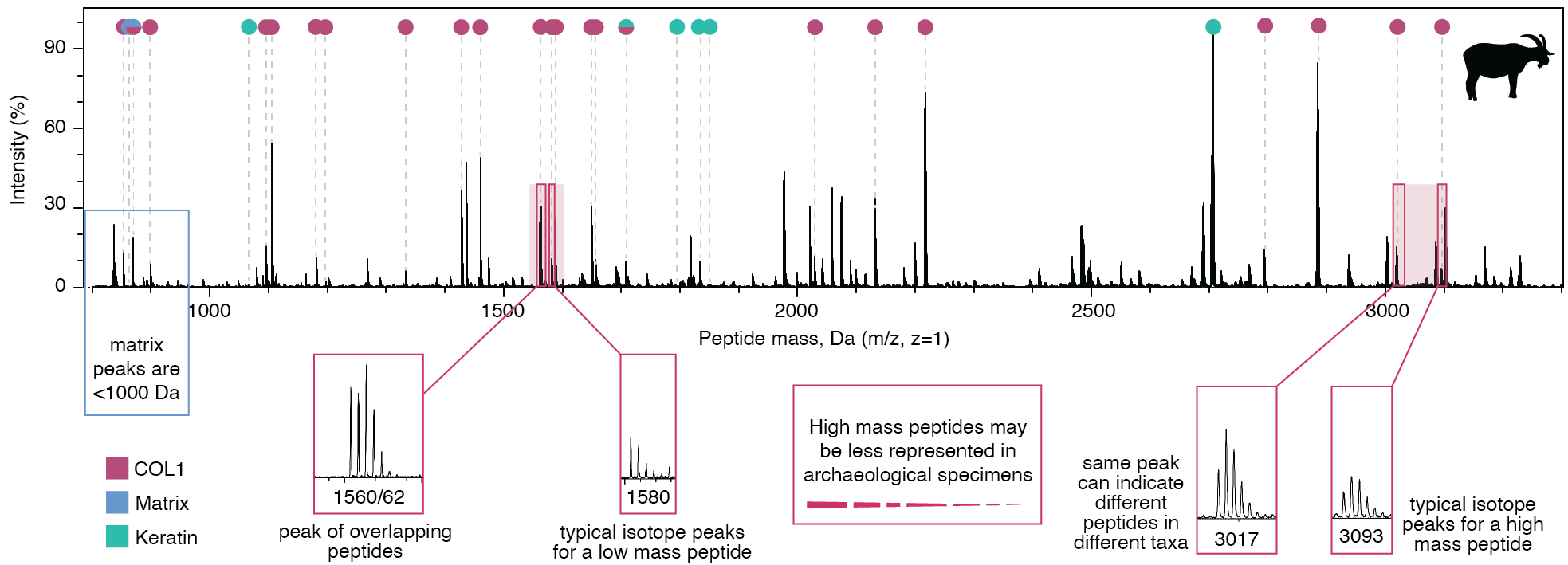 Details of a mass spectrum for goat COL1, with problematic peaks indicated. Peptide isotope distributions differ for low-mass and high-mass peptides. Image adapted from Richter et al. 2022.
Details of a mass spectrum for goat COL1, with problematic peaks indicated. Peptide isotope distributions differ for low-mass and high-mass peptides. Image adapted from Richter et al. 2022.
Marker peptides
Within many collagenous tissues, such as bone, COL1 is the most abundant protein present. ZooMS extraction protocols are optimized to preferentially recover collagen over other proteins, resulting in an extraction product that is overwhelmingly COL1. Prior to analysis by MALDI-TOF mass spectrometry, the collagen is digested with an enzyme, typically trypsin, that cuts the protein at predictable locations to produce smaller peptides. Amino acid sequence differences between taxa result in peptides of different mass that are observed as peaks with different m/z. Using a reference database, it is possible to associate these peaks with specific sequences, thereby allowing taxonomic identification. Peptides that are known to produce good discrimination between taxa are called marker peptides. Prior to becoming marker peptides, candidate peptides are first validated using LC-MS/MS to confirm their amino acid sequence. There are currently 9 mammalian marker peptides in widespread use:
- COL1A1 508-519
- COL1A1 586-618
- COL1A2 292-309
- COL1A2 454-483
- COL1A2 484-498
- COL1A2 502-519
- COL1A2 757-789
- COL1A2 793-816
- COL1A2 978-990
Below is an example of how domestic goats (Capra hircus) and sheep (Ovis aries) can be distinguished from wild springbok (Antidorcas marsupialis) using the COL1A2 502-519 marker peptide. Springbok has a threonine (T) where sheep and goat have an alanine (A), resulting in the springbok COL1A2 502-519 marker peptide having a 30 Da lower m/z.
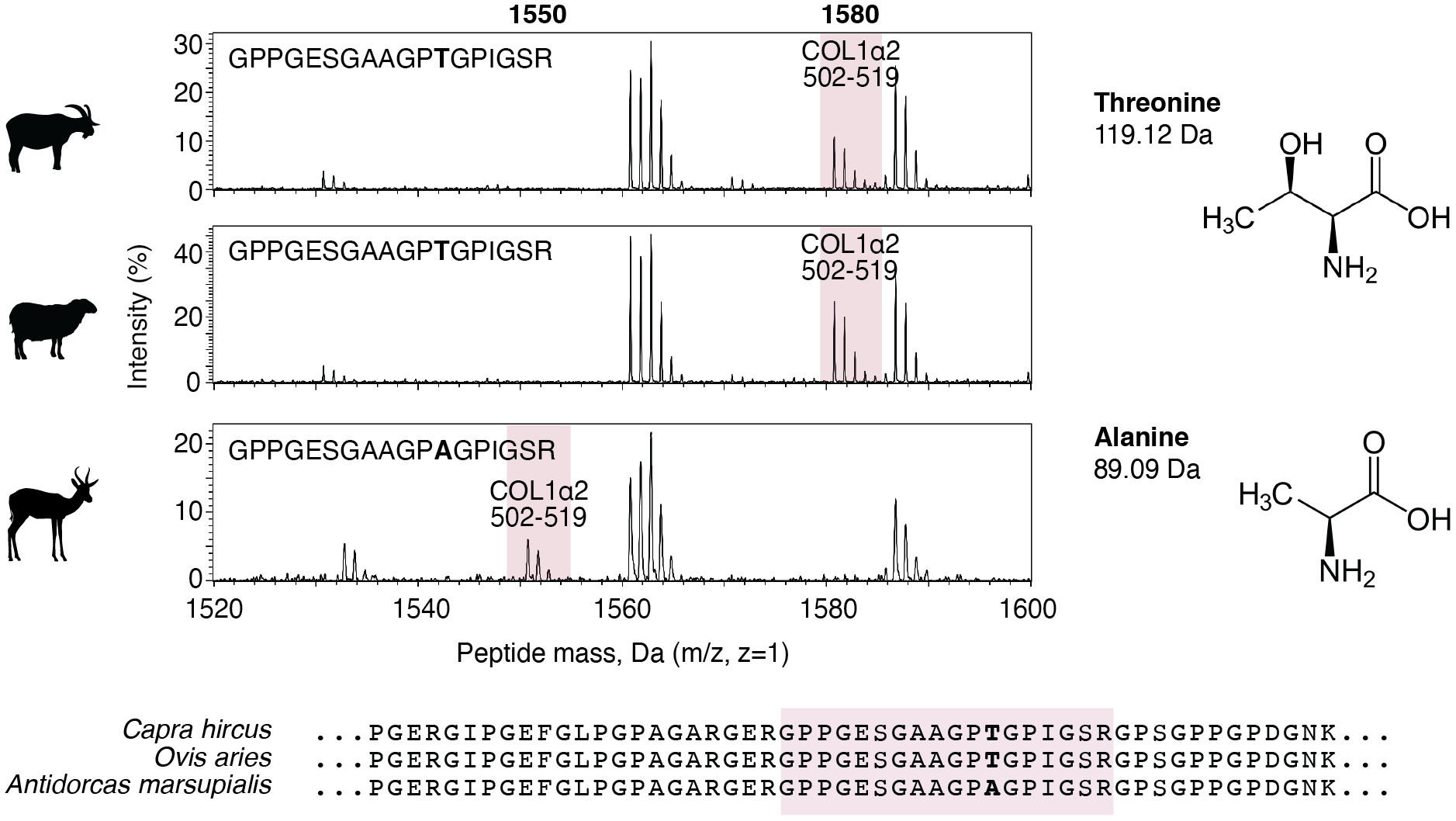 Comparison of peaks in the range 1520-1600 m/z in the collagen mass spectra for goats, sheep, and springbok. The COL1A2 502-519 marker peptide is highlighted in pink, and its corresponding sequence is shown with differences marked in bold. Alanine lacks a COH present in threonine, giving it a 30 Da lower molecular mass. The position of the 502-519 marker peptide (pink) is shown in an alignment of COL1A2 sequences. Image adapted from Richter et al. 2022.
Comparison of peaks in the range 1520-1600 m/z in the collagen mass spectra for goats, sheep, and springbok. The COL1A2 502-519 marker peptide is highlighted in pink, and its corresponding sequence is shown with differences marked in bold. Alanine lacks a COH present in threonine, giving it a 30 Da lower molecular mass. The position of the 502-519 marker peptide (pink) is shown in an alignment of COL1A2 sequences. Image adapted from Richter et al. 2022.
Nomenclature
Several different systems of nomenclature have been used to refer to specific collagen peptides in the ZooMS literature. This can be confusing for beginners. These include systems based on letters (Buckley et al. 2009; Buckley et al. 2014), tryptic position (Richter et al. 2020), and peptide amino acid position (Brown et al. 2021). We recommend using the system of nomenclature based on peptide amino acid position because it can be applied consistenly and systematically across animal taxa.
A schematic showing the numbering system is shown below. The full collagen protein consists of four parts: (1) an N-terminal signal peptide that directs the protein for secretion, (2) a pair of flanking propeptides, (3) a pair of flanking telopeptides, and (4) a central helical protein. After expression, COL1a chains undergo substantial intracellular posttranslational modification and trimming before being secreted as mature helical protein. During this process, the signal peptide and propeptides are removed; they are therefore never found in collagenous tissues. The mature COL1 protein consists of a helical region flanked by short telopeptides. The telopeptides are of varying length between taxa, but the helical region of vertebrate COL1a chains is length invariant, allowing a consistent numbering scheme to be developed. ZooMS nomenclature thus sets position 1 as the start of the helical region, as shown below:
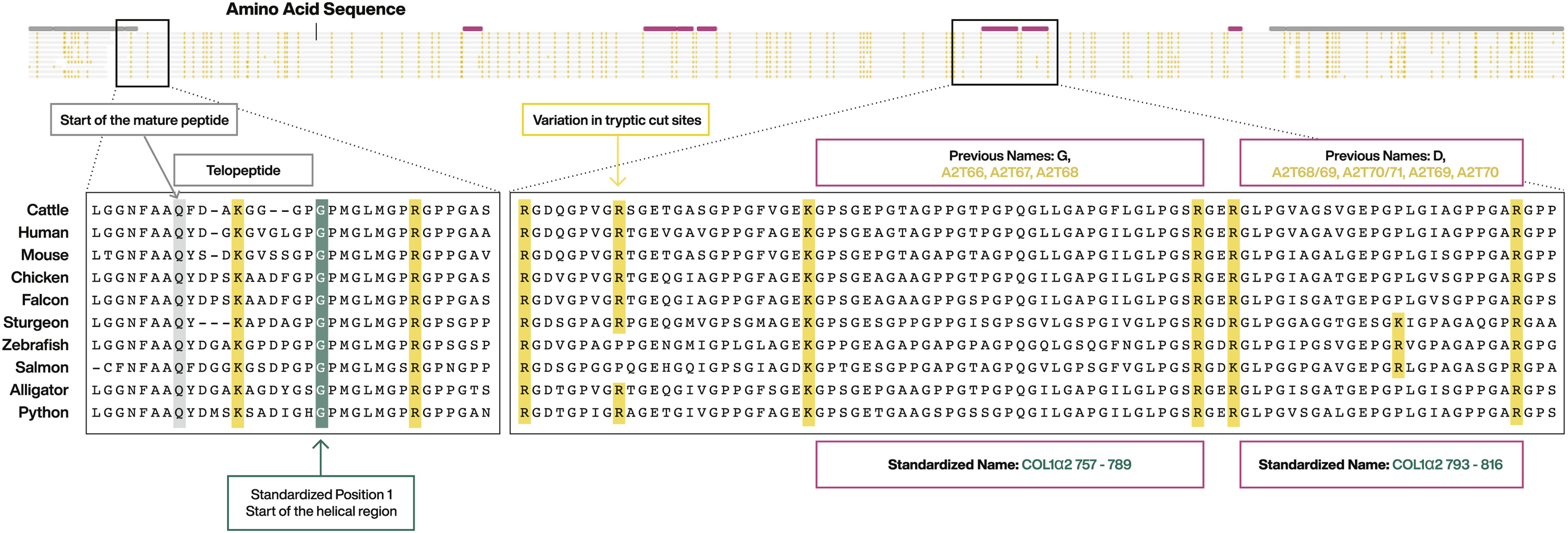 Structure of the COL1 protein. Signal peptide is not shown. Propeptides are indicated by gray bars. Selected marker peptides are indicated by pink bars. Enlarged areas of the spectra show the start of the mature peptide, the N-terminal telopeptide, and the start of the helical region. Tryptic cut sites are highlighted in yellow. Image from Brown et al. 2021.
Structure of the COL1 protein. Signal peptide is not shown. Propeptides are indicated by gray bars. Selected marker peptides are indicated by pink bars. Enlarged areas of the spectra show the start of the mature peptide, the N-terminal telopeptide, and the start of the helical region. Tryptic cut sites are highlighted in yellow. Image from Brown et al. 2021.
For further information about changes in nomenclature through time and for advice on interconverting between systems, see Brown et al. 2021.